Chromatin replication and epigenome maintenance in the Groth Group
The focus of the Groth group is to elucidate how chromatin organization and function is propagated during mitotic cell division to maintain genome and epigenome stability.

Our goal is to understand how mechanisms that underpin epigenetic cell memory impact on cell fate decision in development, cellular reprogramming and epigenetic changes that are prolific in cancer.
Elucidating how chromatin is replicated and propagated across cell division is a major challenge in biology critical to understand development and disease. To move the boundaries of our current understanding, it is essential to identify the molecular mechanisms that orchestrate the interplay between epigenome and genome maintenance. In our group, we take a collaborative and interdisciplinary approach to break new grounds in this field.
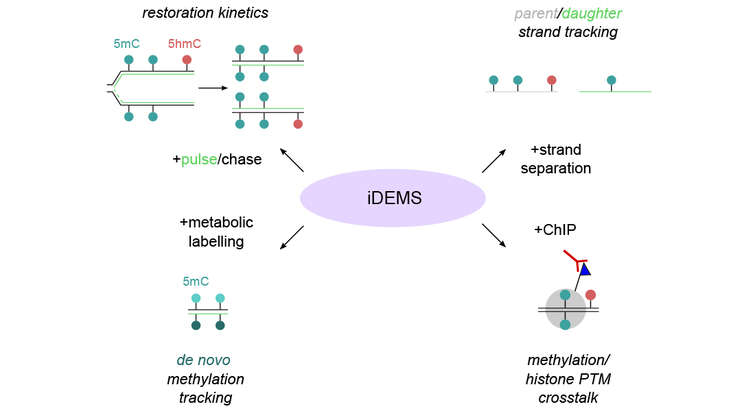
We have developed an array of tailored genomics (ChOR-seq, SCAR-seq, Repli-ATAC) and proteomics technologies (NCC-SILAC) to address chromatin replication and epigenome maintenance. We also apply structure-function studies, chromatin biochemistry, high content imaging and cutting-edge genome editing to dissect chromatin function in DNA repair and protein-based cell memory mechanisms.
Our work has transformed the view of chromatin replication and its implication for epigenome and genome maintenance. We now know that transmission of histone-based information is handled by the replication machinery itself, directly linking epigenetic and genetic inheritance. Surprising, new histones provide a signature for cells to identify sister chromatids and direct error-free repair of DNA lesions through a novel type of histone readers. This mechanism is now being explored for drug discovery to develop DNA repair inhibitors for cancer therapy in the spin-out company, Ankrin Therapeutics (http://ankrin.com/).
Proteome dynamics at broken replication forks reveal a distinct ATM-directed repair response suppressing double-strand break ubiquitination.
Nakamura K, et al. (2021), Molecular Cell
Applying comprehensive proteomics to profile chromatin environment of damaged replication forks, we provide a new framework to understand how cells orchestrate homologous recombination repair of replication-associated DNA double-strand breaks.
Chromatin replication and epigenetic cell memory
Review article in Nature Cell Biology
MCM2 promotes symmetric inheritance of modified histones during DNA replication
By developing SCAR-seq technology to track inheritance of parental histones to the two arms of the replication fork, we discovered that MCM2, part of the replicative helicase, is recycling histones to the lagging strand.
H4K20me0 marks post-replicative chromatin and recruits the TONSL-MMS22L DNA repair complex
Taking a structure-function approach, we identified a new histone reader domain that recruits of homologous recombination proteins TONSL-MMS22L and BARD1-BRCA1 (Nakamura et al., 2019, Nat Cell Biol) to replicated chromatin, licensing error-free DNA repair only when a sister chromatid is available.
Transcription restart establishes chromatin accessibility after DNA replication
Using repli-ATAC-seq, we show that nascent chromatin is inaccessible to the transcription machinery, and that the return of accessibility is heterogenous across genomic features and driven by resumption of transcription post-replication.
Groth group news
Staff list
| Name | Title | Phone | |
|---|---|---|---|
| Anja Groth | Professor, Head of Research | +4535325538 | |
| Kathleen Stewart-Morgan | Visiting Professor | +4535334307 |